Interpretable GNNs for Connectome-Based Brain Disorder Analysis
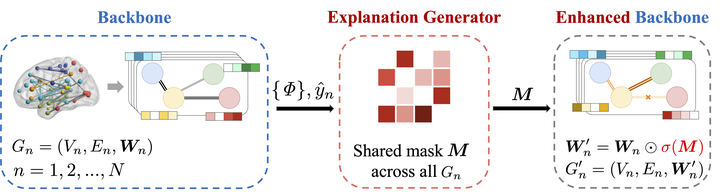 The overview of our proposed interpretable framework: the backbone prediction model is firstly trained on the original data; then the explanation generator is learned based on the trained backbone model; finally we eliminate the noises in the original graph by applying the learned explanation mask and tune the whole enhanced model.
The overview of our proposed interpretable framework: the backbone prediction model is firstly trained on the original data; then the explanation generator is learned based on the trained backbone model; finally we eliminate the noises in the original graph by applying the learned explanation mask and tune the whole enhanced model.Human brains lie at the core of complex neurobiological systems, where the neurons, circuits, and subsystems interact in enigmatic ways. Understanding the structural and functional mechanisms of the brain has long been an intriguing pursuit for neuroscience research and clinical disorder therapy. Mapping the connections of the human brain as a network is one of the most pervasive paradigms in neuroscience. Graph Neural Networks (GNNs) have recently emerged as a potential method for modeling complex network data. Deep models, on the other hand, have low interpretability, which prevents their usage in decision-critical contexts like healthcare. To bridge this gap, we propose an interpretable framework to analyze disorder-specific salient regions of interest and prominent connections. The proposed framework consists of two modules: a brain network-oriented backbone prediction model and a globally shared explanation generator that highlights disorder-specific biomarkers including salient Regions of Interest (ROIs) and important connections. We conduct experiments on three real-world datasets of brain disorders. The results show the outstanding performance and the meaningful biomarkers identified under our framework.